Analysis of CMIP6, ERA5, and CloudSat#
Table of Contents#
1. Introduction #
Questions
How is the cloud phase and snowfall
NOTE: .
2. Data Wrangling #
Organize my data#
Define a prefix for my project (you may need to adjust it for your own usage on your infrastructure).
input folder where all the data used as input to my Jupyter Notebook is stored (and eventually shared)
output folder where all the results to keep are stored
tool folder where all the tools
The ERA5 0.25deg data is located in the folder \scratch\franzihe\, CloudSat at …
import os
import pathlib
import sys
import socket
hostname = socket.gethostname()
abs_path = str(pathlib.Path(hostname).parent.absolute())
WORKDIR = abs_path[:- (len(abs_path.split('/')[-2] + abs_path.split('/')[-1])+1)]
if "mimi" in hostname:
print(hostname)
DATA_DIR = "/mn/vann/franzihe/"
# FIG_DIR = "/uio/kant/geo-geofag-u1/franzihe/Documents/Figures/ERA5/"
FIG_DIR = f"/uio/kant/geo-geofag-u1/franzihe/Documents/Python/globalsnow/CloudSat_ERA5_CMIP6_analysis/Figures/"
elif "glefsekaldt" in hostname:
DATA_DIR = "/home/franzihe/Data/"
FIG_DIR = "/home/franzihe/Documents/Figures/ERA5/"
INPUT_DATA_DIR = os.path.join(DATA_DIR, 'input')
OUTPUT_DATA_DIR = os.path.join(DATA_DIR, 'output')
UTILS_DIR = os.path.join(WORKDIR, 'utils')
FIG_DIR_mci = os.path.join(FIG_DIR, 'McIlhattan/')
sys.path.append(UTILS_DIR)
# make figure directory
try:
os.mkdir(FIG_DIR)
except OSError:
pass
try:
os.mkdir(FIG_DIR_mci)
except OSError:
pass
mimi.uio.no
Import python packages#
Pythonenvironment requirements: file requirements_globalsnow.txtload
pythonpackages from imports.pyload
functionsfrom functions.py
# supress warnings
import warnings
warnings.filterwarnings('ignore') # don't output warnings
# import packages
from imports import(xr, ccrs, cy, plt, glob, cm, fct, np, pd, add_cyclic_point, TwoSlopeNorm, linregress)
# from matplotlib.lines import Line2D
# from matplotlib.patches import Patch
# from sklearn.metrics import r2_score
# plt.rcParams['text.usetex'] = True
from seaborn import set_context
xr.set_options(display_style='html')
<xarray.core.options.set_options at 0x7efc0842c670>
# reload imports
%load_ext autoreload
%autoreload 2
# plot cosmetics
set_context('paper', font_scale=1.5, rc={"lines.linewidth": 2})
Open variables#
Get the data requried for the analysis.
dat_in = os.path.join(OUTPUT_DATA_DIR, 'CS_ERA5_CMIP6')
# make output data directory
# try:
# os.mkdir(dat_out)
# except OSError:
# pass
sic = xr.open_dataset(os.path.join(INPUT_DATA_DIR, 'AMSR2_sea_ice/40NS/AMSR2_IPSL-CM6A-LR_200701_201012.nc'))
ds_cs_2t = xr.open_mfdataset(sorted(glob(os.path.join(INPUT_DATA_DIR, 'cloudsat/aux_monthly_2t/cloudsat*.nc'))))
cs_2t = ds_cs_2t.temp_2m.groupby('time.season').mean('time')
ds_era_2t = xr.open_dataset('/mn/vann/franzihe/output/CS_ERA5_CMIP6/orig/era_500_orig_20070101-20101231.nc')
era_2t = ds_era_2t.tas.groupby('time.season').mean('time')
# Plotting with different markers and colors for each season
colors = cm.hawaii(range(0, 256, int(256 / 4) + 1))
season_colors = {
"DJF": colors[0,:],
"MAM": colors[1,:],
"JJA": colors[2,:],
"SON": colors[3,:]
}
season_markers = {
"DJF": "o", # Circle
"MAM": "s", # Square
"JJA": "D", # Diamond
"SON": "^" # Triangle
}
# fig, ax = plt.subplots(figsize=(10, 6))
# f, axsm = plt.subplots(nrows=1, ncols=2, sharex=True, sharey=True, figsize=[15, 7.5])
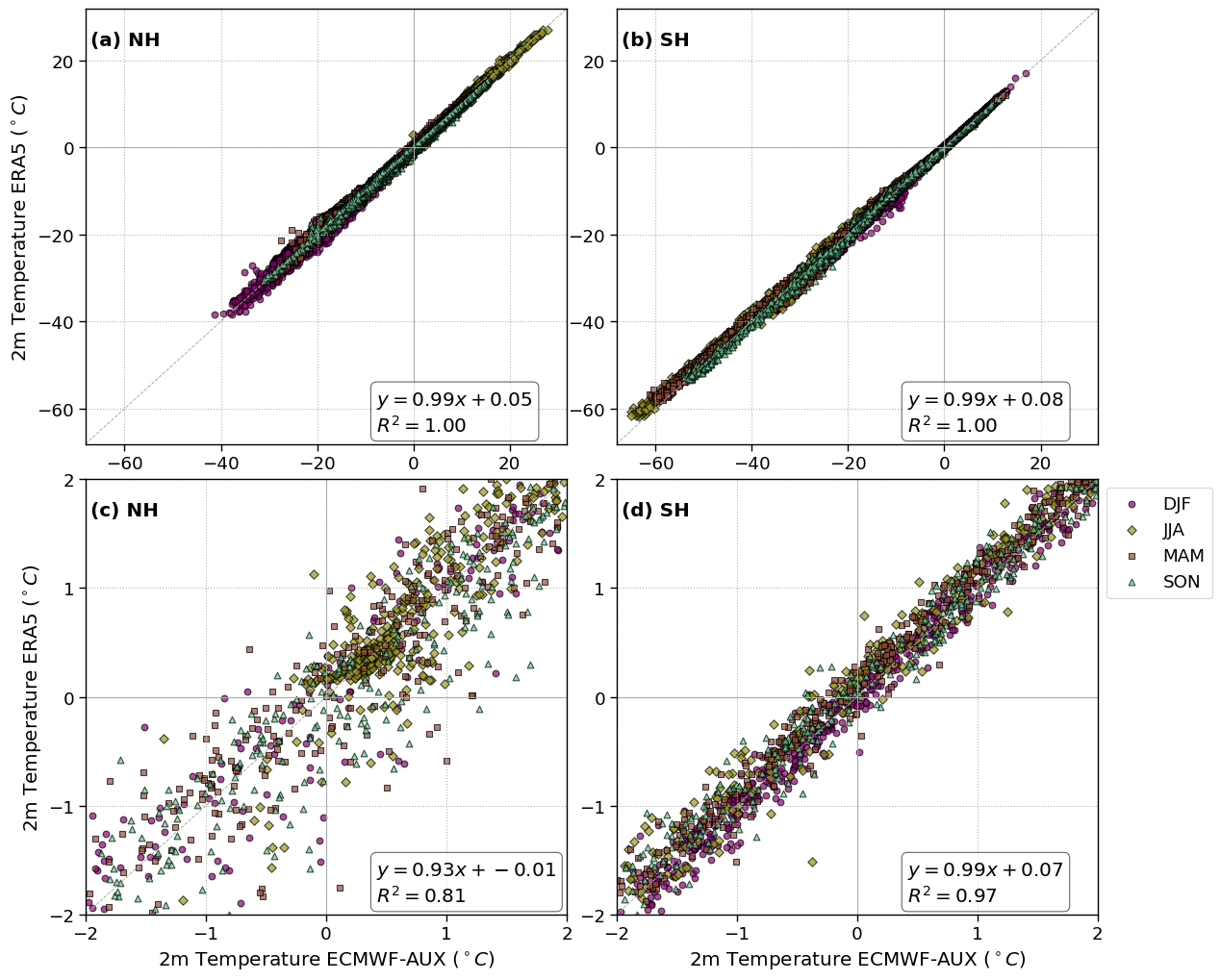
f, axsm = plt.subplots(nrows=2, ncols=2, figsize=[12.5, 10])
for ax, k, hemisphere in zip(axsm.flatten(), range(len(fct.fig_label)), ['NH','SH', 'NH', 'SH']):
ax.grid(linestyle=':')
ax.text(0.01, 0.95, f'{fct.fig_label[k]} {hemisphere}', fontweight='bold', horizontalalignment='left', verticalalignment='top', transform=ax.transAxes)
# ax.set_xlabel('2m Temperature ECMWF-AUX (K)')
ax.axvline(x=273.15-273.15, ymin=0, ymax=1, color='darkgrey', lw=.7)
ax.axhline(y=273.15-273.15, xmin=0, xmax=1, color='darkgrey', lw=.7)
# plot each seasons data
lat_slice = slice(45,90) if hemisphere == 'NH' else slice(-90,-45)
if k == 0 or k ==1:
da_x = (cs_2t-273.15).sel(lat = lat_slice)
da_y = (era_2t-273.15).sel(lat = lat_slice)
# for season in cs_2t.season.values:
# ax.scatter(cs_2t.sel(season=season, lat=lat_slice),
# era_2t.sel(season=season, lat=lat_slice),
# color=season_colors[season],
# marker=season_markers[season],
# label=season,
# alpha=0.7,
# edgecolors="k" # Add black edge for clarity
# )
ax.set_xlim([205-273.15,305-273.15])
ax.set_ylim([205-273.15,305-273.15])
if k == 2 or k == 3:
da_x = (cs_2t-273.15).sel(lat = lat_slice)
da_x = da_x.where((da_x > -2) & (da_x < +2))
da_y = (era_2t-273.15).sel(lat = lat_slice)
da_y = da_y.where((da_y > -2) & (da_y < +2))
ax.set_xlim([-2, +2])
ax.set_ylim([-2, +2])
ax.set_xticks(np.arange(-2,276-273.15))
ax.set_yticks(np.arange(-2,276-273.15))
# Remove NaN values in both x and y
mask = ~np.isnan(da_x.values.ravel()) & ~np.isnan(da_y.values.ravel())
x_clean = da_x.values.ravel()[mask]
y_clean = da_y.values.ravel()[mask]
# Perform polyfit on cleaned data
slope, intercept, r_value, p_value, std_err = linregress(x_clean, y_clean)
# print(f"Slope: {slope}, Intercept: {intercept}, Std err: {std_err}, R value: {r_value}")
for season in cs_2t.season.values:
ax.scatter(da_x.sel(season=season),
da_y.sel(season=season),
color=season_colors[season],
marker=season_markers[season],
label=season,
alpha=0.7,
edgecolors="k" # Add black edge for clarity)
)
ax.plot([0, 1], [0, 1], ls='--', color='darkgrey', lw=.7,transform=ax.transAxes,)
textstr = '\n'.join((
r'$y=%.2f x + %.2f$' % (slope, intercept),
r'$R^2=%.2f$' % (r_value**2, ),))
# these are matplotlib.patch.Patch properties
props = dict(boxstyle='round', facecolor='white', alpha=0.6)#0.5)
# place a text box in upper left in axes coords
ax.text(0.605, 0.125, textstr, transform=ax.transAxes, #fontsize=14,
verticalalignment='top', bbox=props)
axsm[0,0].set_ylabel('2m Temperature ERA5 ($^\circ C$)')
axsm[1,0].set_ylabel('2m Temperature ERA5 ($^\circ C$)')
axsm[1,0].set_xlabel('2m Temperature ECMWF-AUX ($^\circ C$)')
axsm[1,1].set_xlabel('2m Temperature ECMWF-AUX ($^\circ C$)')
axsm[1,1].legend(#loc='lower right',
bbox_to_anchor=(1., 1.0))
plt.tight_layout(pad=0., w_pad=0., h_pad=0.) ;
figname = f'CC_vs_ERA5_temp_NH_SH.png'
plt.savefig(FIG_DIR + figname, format='png', bbox_inches='tight', transparent=True)
ds_cmip_2t = xr.open_dataset('/mn/vann/franzihe/output/CS_ERA5_CMIP6/orig/cmip_500_orig_20070101-20101231.nc')
cmip_2t = ds_cmip_2t.tas.groupby('time.season').mean(('time','model'))
lwp_thresholds = [3, 5, 10, 15,]# 20]
dict_label = {
# 'lcc_wo_snow': {'cb_label':'FsLCC (%)', 'levels':np.arange(0,110,10), 'vmin': 0, 'vmax':100, 'diff_levels':np.arange(-30,35,5), 'diff_vmin':-30, 'diff_vmax':30},
# 'lcc_w_snow': {'cb_label':'FoS in sLCCs (%)', 'levels':np.arange(0,110,10), 'vmin': 0, 'vmax':100, 'diff_levels':np.arange(-60,65,5), 'diff_vmin':-60, 'diff_vmax':60},
# 'sf_eff': {'cb_label':'SE in sLCCs (h$^{-1}$)', 'levels':np.arange(0,5.5,.5), 'vmin':0, 'vmax':5, 'diff_levels':np.arange(-1.2,1.4,.2), 'diff_vmin':-1.2, 'diff_vmax':1.2}#'Relative snowfall efficiency (h$^{-1}$)'
'FLCC' : {'cb_label':'f$_{{LCC}}$ (%)', 'levels':np.arange(0,110.,10.), 'vmin':0, 'vmax': 100., 'vmax2': 100., 'diff_levels':np.arange(-100,110,10), 'diff_vmin':-100, 'diff_vmax':100, 'bounds':np.linspace(-40,40,17), 'qrates': list(np.arange(-42.5, 42.5,5))},
'FsLCC': {'cb_label':'f$_{{sLCC}}$ (%)', 'levels':np.arange(0,110.,10.), 'vmin':0, 'vmax': 100, 'vmax2': 65., 'diff_levels':np.arange(-100,110,10), 'diff_vmin':-100, 'diff_vmax':100, 'bounds':np.arange(-25, 20, 5), 'qrates': list(np.arange(-27.5, 20, 5))},
# 'FoP' : {'cb_label':'FoP in LCCs (%)', 'levels':np.arange(0,105.,5.), 'vmin':0, 'vmax': 100, 'diff_levels':np.arange(-100,110,10), 'diff_vmin':-100, 'diff_vmax':100},
'FoS' : {'cb_label':'f$_{{snow}}$ (%)', 'levels':np.arange(0,110.,10.), 'vmin':0, 'vmax': 100, 'vmax2': 100., 'diff_levels':np.arange(-100,110,10), 'diff_vmin':-100, 'diff_vmax':100, 'bounds':np.arange(-80, 20, 10), 'qrates':list(np.arange(-85, 25, 10))},
# 'pr_eff': {'cb_label':'PE in sLCCs (h$^{-1}$)', 'levels':np.arange(0,550.,50.), 'vmin':0, 'vmax':500, 'diff_levels':np.arange(-120,140,20), 'diff_vmin':-120, 'diff_vmax':120},
'FLCC-FsLCC': {'cb_label':'f$_{{LCC}}$ (%), f$_{{sLCC}}$ (%)', 'levels':np.arange(0,110.,10.), 'vmin':0, 'vmax': 100, 'vmax2': 65., 'diff_levels':np.arange(-100,110,10), 'diff_vmin':-100, 'diff_vmax':100, 'bounds':np.linspace(-45,45,19), 'qrates': list(np.arange(-47.5,47.5,5))},
'sf_eff': {'cb_label':'SE in sLCCs (h$^{-1}$)','levels':np.arange(0,5.5,.5), 'vmin':0, 'vmax': 5, 'vmax2': 7.5, 'diff_levels':np.arange(-1.2,1.4,.2), 'diff_vmin':-1.2, 'diff_vmax':1.2, 'bounds':np.linspace(-2,2,21), 'qrates': list(np.arange(-2.1,2.1,0.2))},
# 'sf_eff': {'cb_label':'SE in sLCCs (h$^{-1}$)','levels':np.arange(0,9.5,.5), 'vmin':0, 'vmax': 9, 'diff_levels':np.arange(-1.2,1.4,.2), 'diff_vmin':-1.2, 'diff_vmax':1.2},
}
# Initialize dictionaries to store data and regression results
ratios = {}
regression = {}
# Loop through different LWP thresholds
for lwp_threshold in lwp_thresholds:
# Load data into ratios dictionary
ratios[lwp_threshold] = xr.open_mfdataset(glob(f'{dat_in}/ratios_500/*LWP{lwp_threshold}_*.nc'))
# Initialize a dictionary to store linear regression results
linregress_results = {}
# Perform linear regression for each variable
for var_name in dict_label.keys():
linregress_results[var_name] = fct.get_linear_regression_hemisphere(ratios[lwp_threshold], var_name)
# Concatenate regression results along the 'variable' dimension
regression[lwp_threshold] = xr.concat(objs=list(linregress_results.values()),
dim =list(linregress_results.keys())).rename({"concat_dim":"variable"})
# Reindex the data according to the desired model order
regression[lwp_threshold] = regression[lwp_threshold].reindex({'model':['ERA5', 'MIROC6', 'CanESM5', 'AWI-ESM-1-1-LR',
'MPI-ESM1-2-LR', 'UKESM1-0-LL', 'HadGEM3-GC31-LL', 'CNRM-CM6-1',
'CNRM-ESM2-1', 'IPSL-CM6A-LR', 'IPSL-CM5A2-INCA']})
# Concatenate data and regression results along the 'threshold' dimension
ratios = xr.concat(objs=list(ratios.values()), dim=lwp_thresholds).rename({"concat_dim":"threshold"})
ratios = ratios.reindex(season=['DJF', 'MAM', 'JJA', 'SON'])
regression = xr.concat(objs=list(regression.values()), dim=lwp_thresholds).rename({"concat_dim":"threshold"})
# Initialize dictionaries to store data and regression results
ratios_mci = {}
regression_mci = {}
# Loop through different LWP thresholds
for lwp_threshold in lwp_thresholds:
# Load data into ratios dictionary
ratios_mci[lwp_threshold] = xr.open_mfdataset(glob(f'{dat_in}/ratios_500_mci/*LWP{lwp_threshold}_*.nc'))
# Initialize a dictionary to store linear regression results
linregress_results = {}
# Perform linear regression for each variable
for var_name in dict_label.keys():
linregress_results[var_name] = fct.get_linear_regression_hemisphere(ratios_mci[lwp_threshold], var_name)
# Concatenate regression results along the 'variable' dimension
regression_mci[lwp_threshold] = xr.concat(objs=list(linregress_results.values()),
dim =list(linregress_results.keys())).rename({"concat_dim":"variable"})
# Reindex the data according to the desired model order
regression_mci[lwp_threshold] = regression_mci[lwp_threshold].reindex({'model':['ERA5', 'MIROC6', 'CanESM5', 'AWI-ESM-1-1-LR',
'MPI-ESM1-2-LR', 'UKESM1-0-LL', 'HadGEM3-GC31-LL', 'CNRM-CM6-1',
'CNRM-ESM2-1', 'IPSL-CM6A-LR', 'IPSL-CM5A2-INCA']})
# Concatenate data and regression results along the 'threshold' dimension
ratios_mci = xr.concat(objs=list(ratios_mci.values()), dim=lwp_thresholds).rename({"concat_dim":"threshold"})
ratios_mci = ratios_mci.reindex(season=['ASO', 'NDJ', 'FMA', 'MJJ'])
regression_mci = xr.concat(objs=list(regression_mci.values()), dim=lwp_thresholds).rename({"concat_dim":"threshold"})
# ratios = xr.open_mfdataset(glob(f'{dat_in}/ratios_500/*LWP{lwp_threshold}_*.nc'))
# ratios_mci = xr.open_mfdataset(glob(f'{dat_in}/ratios_500_mci/*LWP{lwp_threshold}_*.nc'))
# # calculate linear regression and create dataset with values
# linregress = dict()
# for var_name in (dict_label.keys()):
# linregress[var_name] = fct.get_linear_regression_hemisphere(ratios.sel(threshold=lwp_threshold), var_name)
# _ds = list(linregress.values())
# _coord = list(linregress.keys())
# regression = xr.concat(objs=_ds, dim=_coord).rename({"concat_dim":"variable"})
# regression = regression.reindex({'model':['ERA5', 'MIROC6', 'CanESM5', 'AWI-ESM-1-1-LR',
# 'MPI-ESM1-2-LR', 'UKESM1-0-LL', 'HadGEM3-GC31-LL', 'CNRM-CM6-1',
# 'CNRM-ESM2-1', 'IPSL-CM6A-LR', 'IPSL-CM5A2-INCA']})
# # calculate linear regression and create dataset with values
# linregress_mci = dict()
# for var_name in (dict_label.keys()):
# linregress_mci[var_name] = fct.get_linear_regression_hemisphere(ratios_mci, var_name)
# _ds = list(linregress_mci.values())
# _coord = list(linregress_mci.keys())
# regression_mci = xr.concat(objs=_ds, dim=_coord).rename({"concat_dim":"variable"})
# regression_mci = regression_mci.reindex({'model':['ERA5', 'MIROC6', 'CanESM5', 'AWI-ESM-1-1-LR',
# 'MPI-ESM1-2-LR', 'UKESM1-0-LL', 'HadGEM3-GC31-LL', 'CNRM-CM6-1',
# 'CNRM-ESM2-1', 'IPSL-CM6A-LR', 'IPSL-CM5A2-INCA']})
# .sel(model = ['MIROC6', 'CanESM5', 'AWI-ESM-1-1-LR',
# 'MPI-ESM1-2-LR', 'UKESM1-0-LL', 'HadGEM3-GC31-LL', 'CNRM-CM6-1',
# 'CNRM-ESM2-1', 'IPSL-CM6A-LR', 'IPSL-CM5A2-INCA'])
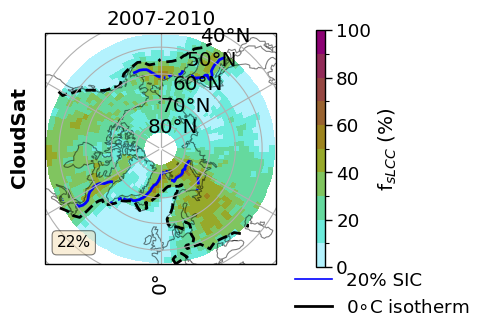
hemisphere = 'NH'
lat_extent = 45.
value = ratios.sel(threshold=lwp_threshold)['FsLCC_season'].sel(model='CloudSat', season='DJF')
value_sic = sic['z_season_mean'].sel( season='DJF')
temperature_da = cs_2t.sel(season='DJF')
hemi_glob = ratios.sel(threshold=lwp_threshold)['FsLCC_season_mean'].sel(model='CloudSat', season='DJF')
projection = fct.create_projection(hemisphere)
f, axsm = plt.subplots(nrows=1,
ncols=1,
subplot_kw={'projection': projection},
figsize=[4, 3], sharex=True, sharey=True)
fct.setup_axes(axsm, hemisphere, lat_extent)
cmap = cm.hawaii_r
levels = dict_label['FoS']['levels']
norm = fct.BoundaryNorm(levels, ncolors=cmap.N, clip=False)
axsm.text(-0.07, 0.55, 'CloudSat',
va='bottom',
ha='center',
rotation='vertical',
rotation_mode='anchor',
transform=axsm.transAxes,
fontweight='bold')
val = value.sel(lat=slice(45,90)) if hemisphere == 'NH' else value.sel(lat=slice(-90,-45))
cf = axsm.pcolormesh(val.lon, val.lat, (val.where(~np.isnan(val))),
transform=ccrs.PlateCarree(),
cmap=cmap,
norm=norm)
contour = value_sic.plot.contour(ax=axsm, transform=ccrs.PlateCarree(), x='lon', y='lat', levels = [20., ], lw=1.2, add_colorbar=False, colors = 'blue')
isotherm = axsm.contour(
temperature_da.lon,
temperature_da.lat,
temperature_da,
transform=ccrs.PlateCarree(),
levels=[273.15],
colors="k", # Color for the isotherm line
linewidths=2,
linestyles="--" # Dashed line for visibility
)
fct.add_text_box(axsm, hemi_glob.sel(hemisphere=hemisphere,), 'FoS')
axsm.set_title('2007-2010')
cbaxes = f.add_axes([0.9, 0.1, 0.0225, 0.79])
cb_label = dict_label['FsLCC']['cb_label']
extend = None
plt.colorbar(cf, cax=cbaxes, shrink=0.5,extend=extend, orientation='vertical', label=cb_label)
legend_elements = [fct.Line2D([0], [0], color='b', lw=1.2, label='20% SIC'),
fct.Line2D([0], [1], color='k', lw=2., label='0$\circ$C isotherm')
]
leg = axsm.legend(handles=legend_elements, bbox_to_anchor=(1.05, 0.0), loc=2, borderaxespad=0.)#loc='best')
leg.get_frame().set_edgecolor('b')
leg.get_frame().set_linewidth(0.0)
# axsm.clabel(contour, fmt='%1.1f', fontsize=12)# cbar = plt.colorbar(contour, )#cax=axsm, orientation='vertical',pad=0.05, shrink = 0.8)
#cbar.set_label('Sea ice concentration (%)',fontsize=16)
# ratios.sel(threshold=lwp_threshold)['FoS_year'].sel(model='ERA5').plot()
# for var_name in (dict_label.keys()):
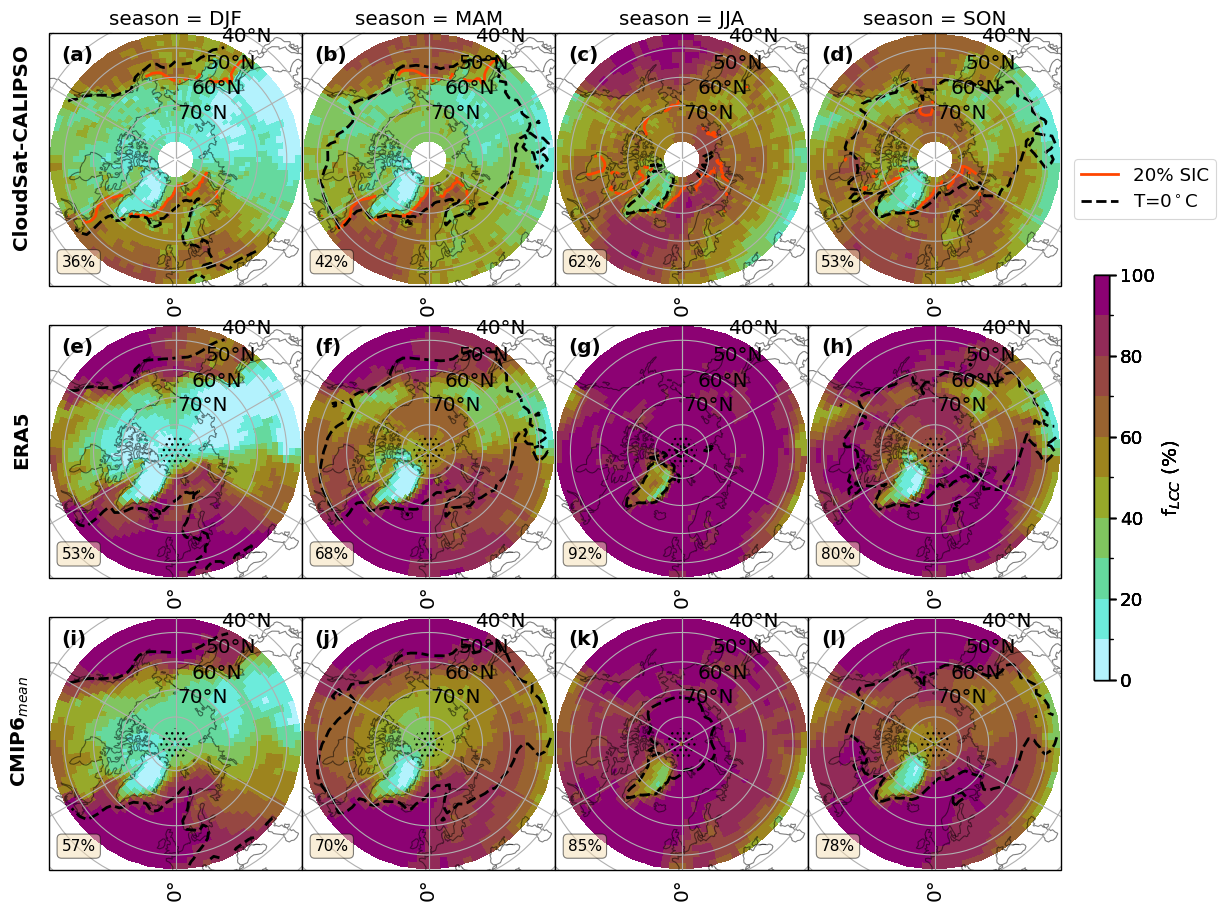
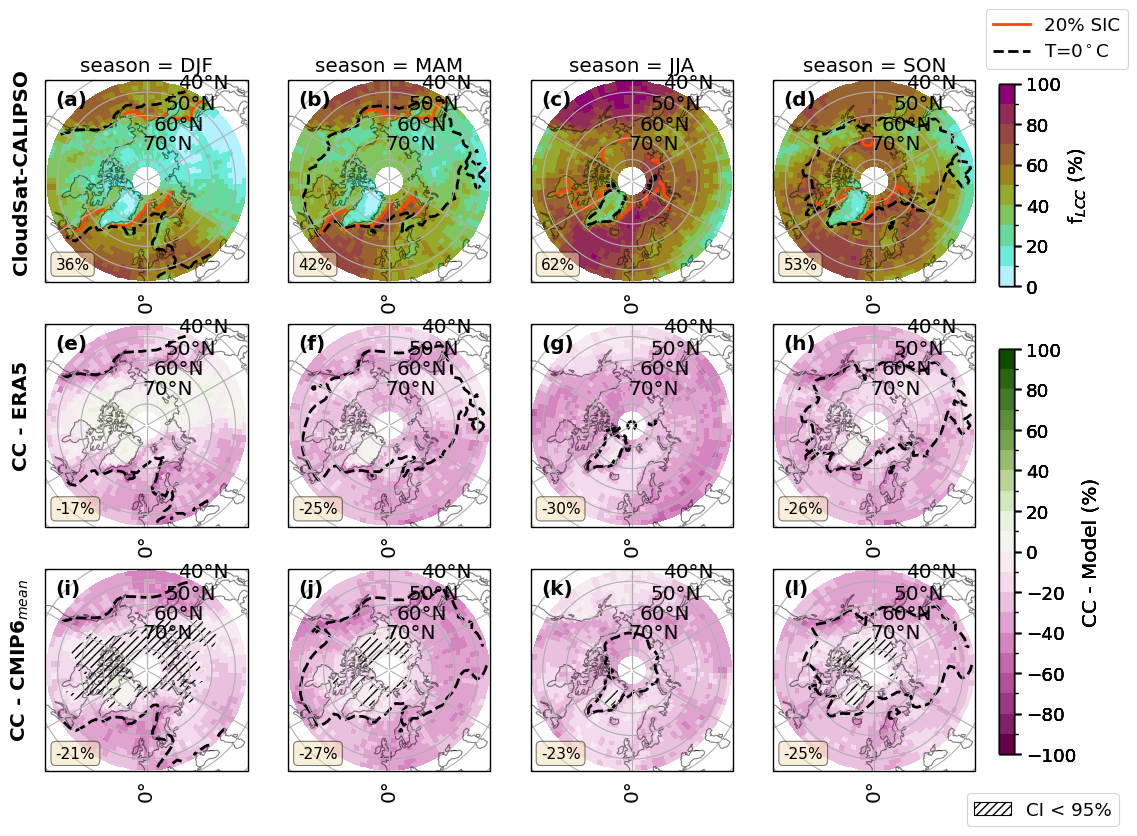
for var_name in ['FLCC']:# 'FLCC','FLCC-FsLCC','sf_eff', 'FsLCC','FoS',]:
# # for lwp_threshold in lwp_thresholds:
for lwp_threshold in [5,]:
# print(f'plot {var_name} for {lwp_threshold}')
F_DIR = os.path.join(FIG_DIR,f'CS_ERA5_CMIP6_{lwp_threshold}/')
F_DIR_mci = os.path.join(FIG_DIR_mci,f'CS_ERA5_CMIP6_{lwp_threshold}/')
try:
os.mkdir(F_DIR)
except OSError:
pass
try:
os.mkdir(F_DIR_mci)
except OSError:
pass
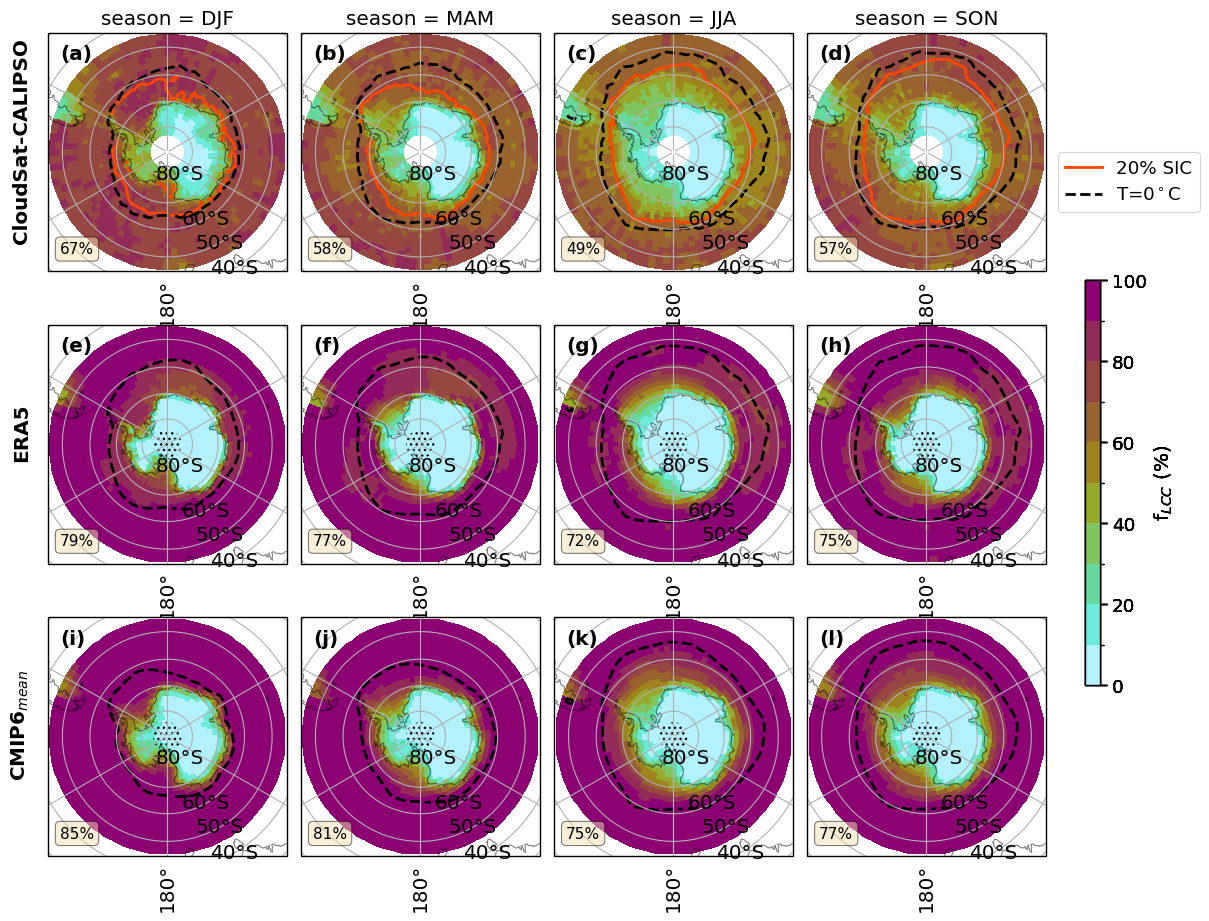
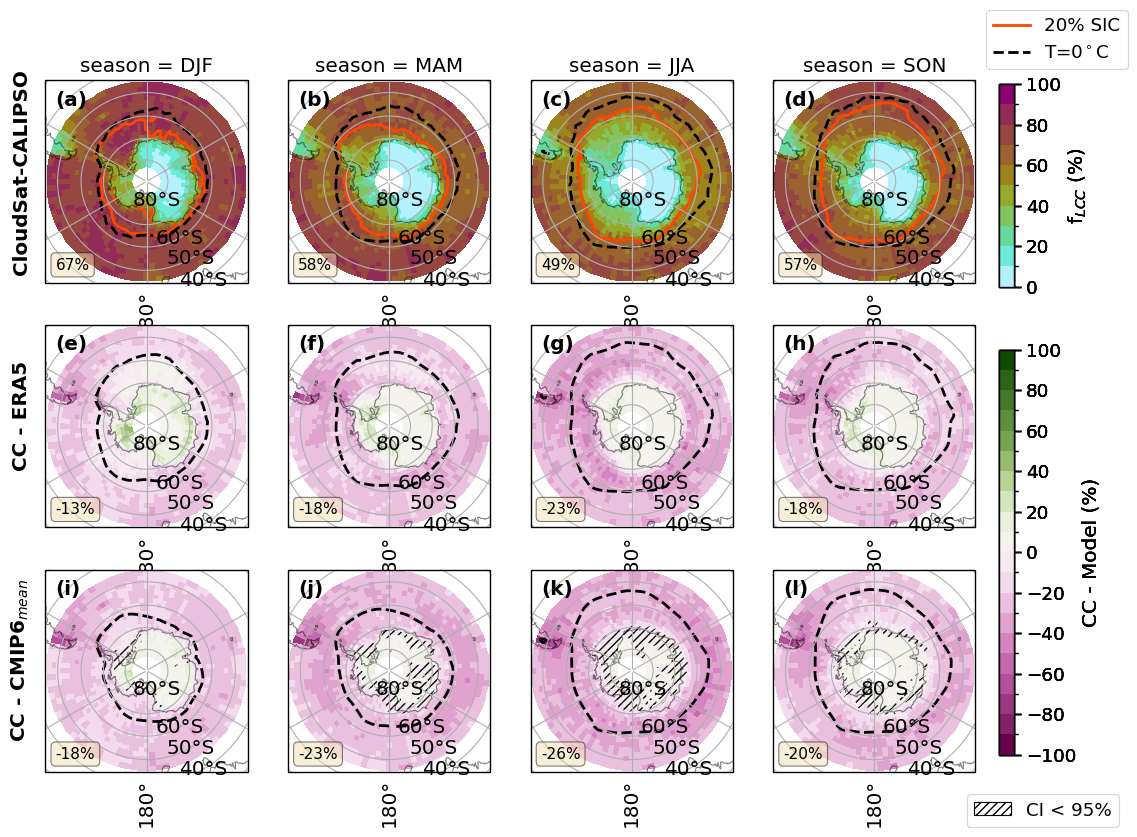
fct.plt_spatial_season_var(ratios.sel(threshold=lwp_threshold), var_name, dict_label, sic['z_season_mean'],cs_2t,era_2t,cmip_2t, F_DIR, 45, lwp_threshold)
# # # fct.plt_spatial_season_var(ratios_mci.sel(threshold=lwp_threshold), var_name, dict_label, F_DIR_mci, 66, lwp_threshold)
# plot monthly model variation
# fct.plt_monthly_model_variation(ratios.sel(threshold=lwp_threshold), var_name, dict_label[var_name],F_DIR, lwp_threshold)
# # # # fct.plt_monthly_model_variation(ratios_mci.sel(threshold=lwp_threshold), var_name, dict_label[var_name],F_DIR_mci, lwp_threshold)
# fct.plt_monthly_interannual_variation(ratios.sel(threshold=lwp_threshold), var_name, lwp_threshold, F_DIR, dict_label[var_name])
# # # # fct.plt_monthly_interannual_variation(ratios_mci.sel(threshold=lwp_threshold), var_name, lwp_threshold, F_DIR_mci, dict_label[var_name])
# # fct.plt_absolute_difference_season(ratios.sel(threshold=lwp_threshold), var_name, lwp_threshold, dict_label[var_name], F_DIR)
# fct.plt_difference_season(ratios.sel(threshold=lwp_threshold), var_name, lwp_threshold, dict_label[var_name], F_DIR)
# # fct.plt_heatmap_all_models_season(ratios.sel(threshold=lwp_threshold), var_name, lwp_threshold, dict_label[var_name], F_DIR)
# # plot scatter CloudSat vs model
# fct.plt_scatter_obs_model(ratios.sel(threshold=lwp_threshold), regression.sel(threshold=lwp_threshold), var_name, dict_label[var_name], F_DIR, lwp_threshold)
# # # fct.plt_scatter_obs_model(ratios_mci.sel(threshold=lwp_threshold), regression_mci.sel(threshold=lwp_threshold), var_name, dict_label[var_name], F_DIR_mci, lwp_threshold)
# # plot variable for individual model
# fct.plt_spatial_season_all_models(ratios.sel(threshold=lwp_threshold), var_name, dict_label, F_DIR, 45)
# # # # fct.plt_spatial_season_all_models(ratios_mci.sel(threshold=lwp_threshold), var_name, dict_label, F_DIR_mci, 66)
# plot difference between CloudSat and ERA5, CMIP6 mean for lwp threshold comparison
# if var_name == 'FLCC' or var_name == 'FsLCC' or var_name == 'FoS':
# for hemisphere in ['NH', 'SH']:
# fct.plt_spatial_annual_difference(ratios, hemisphere, var_name, dict_label[var_name], FIG_DIR, 45, lwp_thresholds)
# fct.plt_spatial_annual_difference(ratios_mci, hemisphere, var_name, dict_label[var_name], F_DIR_mci, 45, lwp_thresholds)
# for lwp_threshold in lwp_thresholds:
# # # plot R2 values for all values and both hemispheres
# F_DIR = os.path.join(FIG_DIR,f'CS_ERA5_CMIP6_{lwp_threshold}/')
# fct.plt_R2_heatmap_season(regression.sel(threshold=lwp_threshold), dict_label, F_DIR, lwp_threshold)
# # # fct.plt_R2_heatmap_season(regression_mci.sel(threshold=lwp_threshold), dict_label, F_DIR_mci, lwp_threshold)
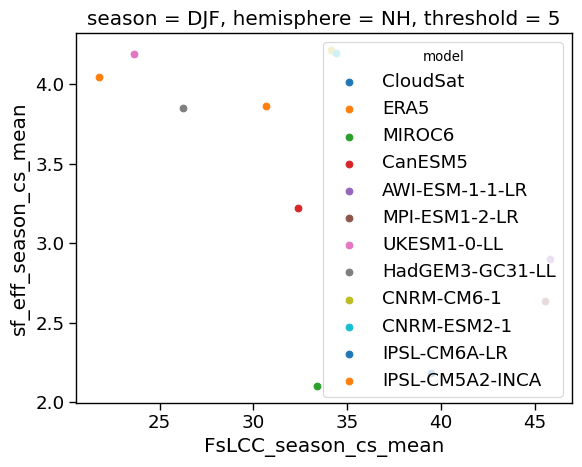
x=ratios['FsLCC'+'_season_cs_mean'].sel(threshold=5, hemisphere='NH', season='DJF')
y=ratios['sf_eff'+'_season_cs_mean'].sel(threshold=5, hemisphere='NH', season='DJF')
xr.plot.scatter(ratios.sel(threshold=5, hemisphere='NH', season='DJF'), x='FsLCC'+'_season_cs_mean', y='sf_eff'+'_season_cs_mean', hue='model', hue_style='discrete', cmap=cm.hawaii)
[<matplotlib.collections.PathCollection at 0x7fd1d81374f0>,
<matplotlib.collections.PathCollection at 0x7fd1d80f5e40>,
<matplotlib.collections.PathCollection at 0x7fd1cbf2d840>,
<matplotlib.collections.PathCollection at 0x7fd1dafe85e0>,
<matplotlib.collections.PathCollection at 0x7fd1d9e12e30>,
<matplotlib.collections.PathCollection at 0x7fd1d9e951e0>,
<matplotlib.collections.PathCollection at 0x7fd1d8127130>,
<matplotlib.collections.PathCollection at 0x7fd1da3b5360>,
<matplotlib.collections.PathCollection at 0x7fd1d80f7670>,
<matplotlib.collections.PathCollection at 0x7fd1da14b250>,
<matplotlib.collections.PathCollection at 0x7fd1d835f9d0>,
<matplotlib.collections.PathCollection at 0x7fd1d9e2c1c0>]
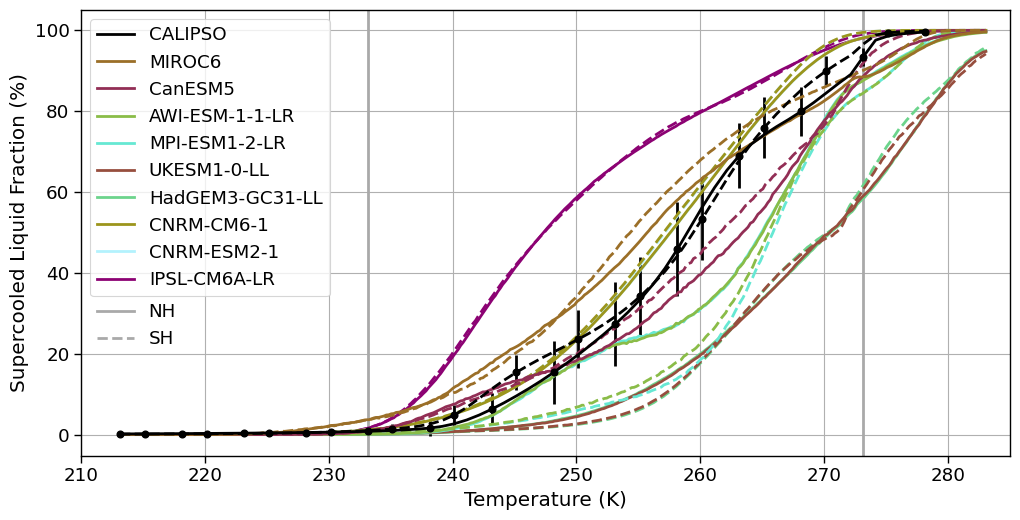
_in2 = os.path.join(INPUT_DATA_DIR, 'cmip6_hist/')
da_sat_NH = pd.read_csv(glob(f'{_in2}/*satellite*NH*')[0], index_col=0)
da_sat_SH = pd.read_csv(glob(f'{_in2}/*satellite*SH*')[0], index_col=0)
da_sat_NH.set_axis(list(np.arange(-60, 6,1)+273.15), axis='columns', inplace=True)
da_sat_SH.set_axis(list(np.arange(-60, 6,1)+273.15), axis=1, inplace=True)
da = pd.read_csv(glob(f'{_in2}/*lcf*')[0])
models = list(da['model'].unique())
da_sat_NH_mean = da_sat_NH.mean(axis=0, skipna=True)
da_sat_SH_mean = da_sat_SH.mean(axis=0, skipna=True)
da_sat_NH_std = da_sat_NH.std(axis=0, skipna=True)
da_sat_SH_std = da_sat_SH.std(axis=0, skipna=True)
f, axsm = plt.subplots(nrows=1, ncols=1, sharex=True, sharey=True, figsize=[10, 5])
# ax = axsm.flat
# colors = cm.hawaii(range(0, 256, int(256 / len(list(da['model'].unique()))) + 1))
cmap = cm.hawaiiS
# for (i, hemisphere) in enumerate(['north', 'south']):
axsm.axvline(x=273.15, ymin=0, ymax=1, color='darkgrey')
axsm.axvline(x=273.15-40, ymin=0, ymax=1, color='darkgrey')
# axsm.text(0.05, 0.95, f'{fct.fig_label[i]}', fontweight='bold', horizontalalignment='left', verticalalignment='top', transform=axsm.transAxes)
axsm.set_xlim([210, 285])
# axsm.fill_between(x=da_sat_NH.columns.values, y1=(da_sat_NH_mean-da_sat_NH_std).values, y2=(da_sat_NH_mean+da_sat_NH_std).values,
# color='k', alpha=0.25)
# axsm.fill_between(x=da_sat_SH.columns.values, y1=(da_sat_SH_mean-da_sat_SH_std).values, y2=(da_sat_SH_mean+da_sat_SH_std).values,
# color='k', alpha=0.25)
axsm.errorbar(x=da_sat_NH_mean.index[::5].values, y=da_sat_NH_mean[da_sat_NH_mean.index[::5]], yerr=da_sat_NH_std[da_sat_NH_std.index[::5]], fmt='o', color ='k', )
axsm.errorbar(x=da_sat_SH_mean.index[2::5].values, y=da_sat_SH_mean[da_sat_SH_mean.index[2::5]], yerr=da_sat_SH_std[da_sat_SH_std.index[2::5]], fmt='o', color ='k', )
# axsm.errorbar(x=da_sat_NH_mean.index.values, y=da_sat_SH_mean, yerr=da_sat_NH_std, errorevery=5, fmt='o', color ='k', )
# plot models
for c, model in zip(plt.cycler('color', cmap.colors), list(da['model'].unique())[::-1]):
da_model = da[da['model'] == model]
da_model[da['region'] =='north'].plot.line(ax=axsm, x='T', y='LCF_weighted_mean', label=model, color=c['color'])
da_model[da['region'] =='south'].plot.line(ax=axsm, x='T', y='LCF_weighted_mean', label='_', color=c['color'], linestyle = '--')
# plot CloudSat
da_sat_NH_mean.plot(ax=axsm, label='CALIPSO', color='k')
da_sat_SH.mean(axis=0, skipna=True).plot(ax=axsm, label='_', color='k', linestyle = '--')
axsm.set_ylabel('Supercooled Liquid Fraction (%)')
axsm.set_xlabel('Temperature (K)')
axsm.set_yticks(np.arange(0, 1.20,.20), np.arange(0, 120,20))
axsm.grid(True)
# Second legend lines
line_solid = fct.Line2D([], [], color='darkgray', linestyle='-', label='NH')
line_dashed = fct.Line2D([], [], color='darkgray', linestyle='--', label='SH')
h, l = axsm.get_legend_handles_labels()
first_legend = axsm.legend(handles=h[::-1], labels=l[::-1], loc=0)
ax = plt.gca().add_artist(first_legend)
second_legend = plt.legend(handles=[line_solid, line_dashed], loc=0, bbox_to_anchor=(-0.38, 0.05, 0.5, 0.33))
second_legend.set_frame_on(False)
plt.tight_layout(pad=0., w_pad=0., h_pad=0.) ;
figname = f'T_vs_LCF_NH_SH.png'
plt.savefig(FIG_DIR + figname, format='png', bbox_inches='tight', transparent=True)
def find_temp_IQR_from_SLF_ratio(da_model, liquid_frac):
# find the index where the model has e.g., 50% ice and 50% liquid in the lower quartile and upper quartile
idx_lower = abs(da_model['LCF_0.25'] - liquid_frac).argmin()
idx_upper = abs(da_model['LCF_0.75'] - liquid_frac).argmin()
# use the index to locate the T5050 at the lower and upper quartile
T_lower_q = da_model['T'].loc[idx_lower]
T_upper_q = da_model['T'].loc[idx_upper]
# IQR temperature variation for this model is the difference between the upper and lower temperature
T = abs(T_upper_q - T_lower_q)
return(T)
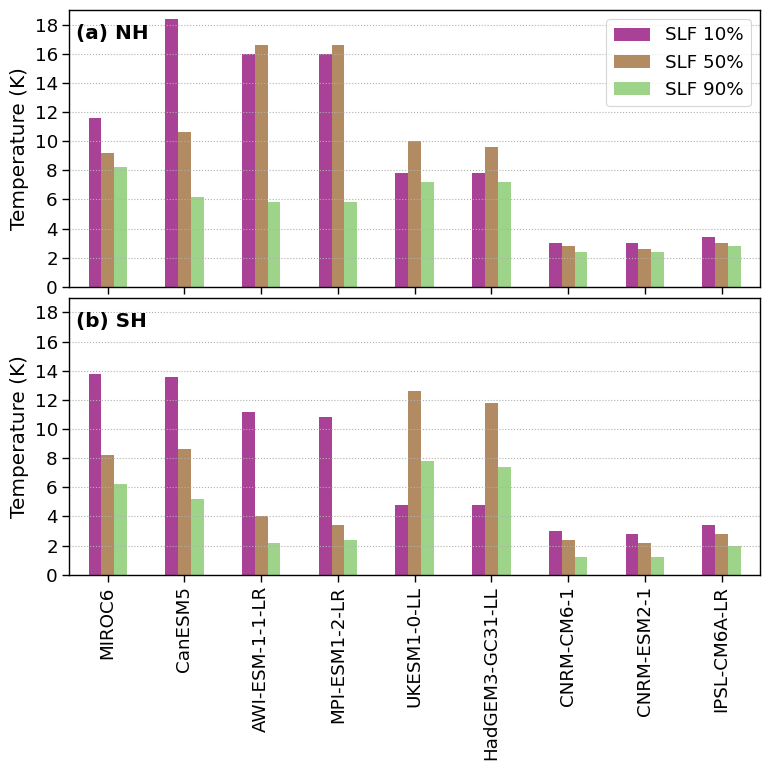
IQR_T_SH = pd.DataFrame(index=models,columns=['SLF 10%', 'SLF 50%', 'SLF 90%'])
IQR_T_NH = pd.DataFrame(index=models,columns=['SLF 10%', 'SLF 50%', 'SLF 90%'])
for model in list(models):
# select the model from the dataset
da_model = da[da['model'] == model]
# select the hemisphere
model_SH = da_model[da['region'] =='south']
model_SH.reset_index(drop=True, inplace=True)
model_NH = da_model[da['region'] =='north']
model_NH.reset_index(drop=True, inplace=True)
IQR_T_SH.loc[model]['SLF 50%'] = find_temp_IQR_from_SLF_ratio(model_SH, 0.5)
IQR_T_SH.loc[model]['SLF 10%'] = find_temp_IQR_from_SLF_ratio(model_SH, 0.1)
IQR_T_SH.loc[model]['SLF 90%'] = find_temp_IQR_from_SLF_ratio(model_SH, 0.9)
IQR_T_NH.loc[model]['SLF 50%'] = find_temp_IQR_from_SLF_ratio(model_NH, 0.5)
IQR_T_NH.loc[model]['SLF 10%'] = find_temp_IQR_from_SLF_ratio(model_NH, 0.1)
IQR_T_NH.loc[model]['SLF 90%'] = find_temp_IQR_from_SLF_ratio(model_NH, 0.9)
f, axsm = plt.subplots(nrows=2, ncols=1, sharex=True, sharey=True, figsize=[7.5, 7.5])
colors = cm.hawaii(range(0, 256, int(256 / 3) + 1))
ax = axsm.flatten()
IQR_T_NH.plot.bar(ax=ax[0], color =colors, alpha=0.75)
IQR_T_SH.plot.bar(ax=ax[1], color =colors, alpha=0.75, legend=False)
for ax, k, hemisphere in zip(axsm.flatten(), range(len(fct.fig_label)), ['NH', 'SH']):
ax.grid(axis='y', linestyle=':')
ax.text(0.01, 0.95, f'{fct.fig_label[k]} {hemisphere}', fontweight='bold', horizontalalignment='left', verticalalignment='top', transform=ax.transAxes)
ax.set_ylabel('Temperature (K)')
ax.set_ylim([0, 19])
ax.set_yticks(np.arange(0,19,2))
plt.tight_layout(pad=0., w_pad=0., h_pad=0.) ;
figname = f'IQR_T_SLF_NH_SH.png'
plt.savefig(FIG_DIR + figname, format='png', bbox_inches='tight', transparent=True)
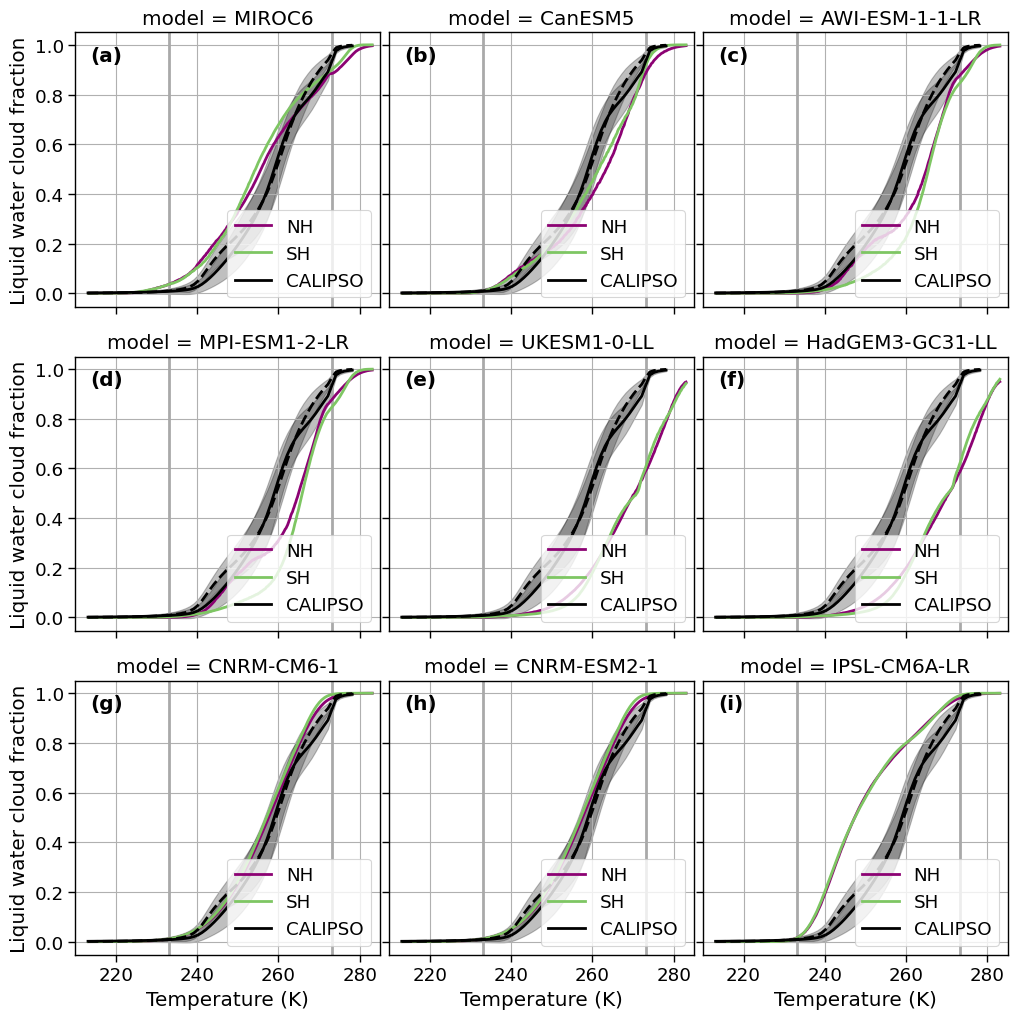
f, axsm = plt.subplots(nrows=3, ncols=3, sharex=True, sharey=True, figsize=[10, 10])
colors = cm.hawaii(range(0, 256, int(256/3)+1))
for ax, model, k in zip(axsm.flatten(), list(da['model'].unique()), range(len(fct.fig_label))):
ax.text(0.05, 0.95, f'{fct.fig_label[k]}', fontweight='bold', horizontalalignment='left', verticalalignment='top', transform=ax.transAxes)
ax.fill_between(x=da_sat_NH.columns.values, y1=(da_sat_NH_mean-da_sat_NH_std).values, y2=(da_sat_NH_mean+da_sat_NH_std).values,
color='k', alpha=0.25)
ax.fill_between(x=da_sat_SH.columns.values, y1=(da_sat_SH_mean-da_sat_SH_std).values, y2=(da_sat_SH_mean+da_sat_SH_std).values,
color='k', alpha=0.25)
ax.axvline(x=273.15, ymin=0, ymax=1, color='darkgrey')
ax.axvline(x=273.15-40, ymin=0, ymax=1, color='darkgrey')
ax.set_xlim([210, 285])
ax.set_title(f'model = {model}')
da_model = da[da['model'] == model]
da_model[da['region'] =='north'].plot.line(ax=ax, x='T', y='LCF_weighted_mean', label='NH', color=colors[0])
da_model[da['region'] =='south'].plot.line(ax=ax, x='T', y='LCF_weighted_mean', label='SH', color=colors[2] )
# plot CloudSat
da_sat_NH_mean.plot(ax=ax, label='CALIPSO', color='k')
da_sat_SH.mean(axis=0, skipna=True).plot(ax=ax, label='_', color='k', linestyle = '--')
ax.set_ylabel('Liquid water cloud fraction')
ax.set_xlabel('Temperature (K)')
ax.grid(True)
ax.legend(loc=4)
plt.tight_layout(pad=0, w_pad=0., h_pad=1) ;
figname = f'T_vs_LCF_models_seperate.png'
plt.savefig(FIG_DIR + figname, format='png', bbox_inches='tight', transparent=True)
# file_pattern = f'{DATA_DIR}/output/CS_ERA5_CMIP6/orig/cloudsat_500_orig*.nc'
# files = sorted(glob(file_pattern))
# weights= xr.open_mfdataset(files)['areacella']
# weights
# dataset = ratios.sel(threshold=5)
# di = dataset[var_name+'_season'] - dataset[var_name+'_season'].sel(model='CloudSat')
# di_2 = np.sqrt(di)
# # di_2_mean = di_2.mean()
# lat_slice = slice(45,90)
# di_2_mean, _, _ = fct.calculate_stats(di_2, weights, lat_slice)
# # dataset[var_name+'_season_cs']
# np.sqrt(di_2_mean)
# for var_name in ['FoS',]:# 'sf_eff', 'FLCC','FLCC-FsLCC','FsLCC',:
# # plot variable for individual model
# fct.plt_spatial_season_all_models(ratios, var_name, dict_label, FIG_DIR, 45)
# fct.plt_spatial_season_all_models(ratios_mci, var_name, dict_label, FIG_DIR_mci, 66)
